MAAB using a fast cycle PCR instrument and a non-labeled probe on LightScanner32
Introduction
The high resolution melting curve can detect changes in gene sequences in the hybrid by scanning amplified fragments of the PCR. In combination with high-resolution saturated fluorescent dyes, the sensitivity of SNPs, insertions or deletions of PCR fragments less than 400 bp can be over 98%. Since its development, this method has been continuously applied to new fields, such as small fragment amplification and unlabeled probe (LunaProbes) for genotyping known sequences. The 3' end of the unlabeled probe is blocked to prevent amplification during PCR, and the difference in melting temperature (Tm value) of the saturated dye LCGreen Plus and unlabeled probe (LunaProbes) of double-stranded DNA is used to distinguish different genes. type. Unlabeled probes can be used to quantify alleles in genotyping, and mutations in alleles with a mutation rate of ≦10% can be detected.
Recently, a method for performing a biased amplification of mutant alleles (MAAB) based on high-resolution melting curve analysis of a non-labeled probe on a fast-circulation real-time PCR instrument and LightScanner32 has been developed. The method can increase the sensitivity of non-labeled probes in detecting allelic mutations.
Recently, a method for performing a biased amplification of mutant alleles (MAAB) based on high-resolution melting curve analysis of a non-labeled probe on a fast-circulation real-time PCR instrument and LightScanner32 has been developed. The method can increase the sensitivity of non-labeled probes in detecting allelic mutations.
Mutant Allele Amplification Bias (MAAB) principle
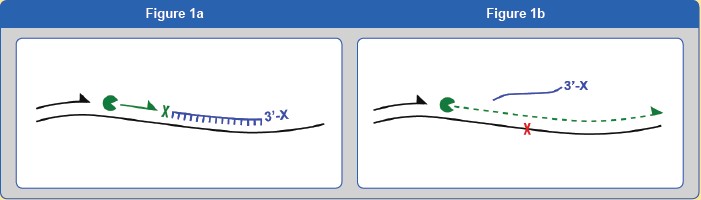
The 3' end of the unlabeled probe is blocked to prevent amplification during PCR. The design of the probe is completely complementary to the wild-type allele, and the allelic site of the mutation is not completely complementary. The biased amplification of the mutated allele is primarily achieved by setting the annealing temperature of the PCR reaction. The annealing temperature of the PCR is lower than the annealing temperature of the wild-type allele probe and higher than the annealing temperature of the mutant allele, and the Tm value and the non-complete complementation of the fully complementary probe (wild-type site) The needle (the site of the mutant) is between the Tm values. At this annealing temperature, the fully complementary probe and the target DNA strand are still bound together (Figure 1a), thus inhibiting the PCR reaction of the wild-type sequence (Figure 1b), thereby achieving a biased amplification of the mutant allele.
method
Unlabeled probes were used to detect mutations in the alleles with a mutation rate of %1% in the context of wild-type alleles, using different annealing temperatures and different exonucleases. Three different genes were selected for detection: TP53 (exon 8) cancer-associated mutations, PAH (exon 11) and PKU (phenylketonuria)-related mutations, and a drug-related therapy for malaria-related genes. The resistant mutation is then subjected to temperature gradient PCR using a conventional hot start PCR instrument to determine the Tm value of the unlabeled probe and whether the normal hot start PCR instrument can complete the MAAB.
result
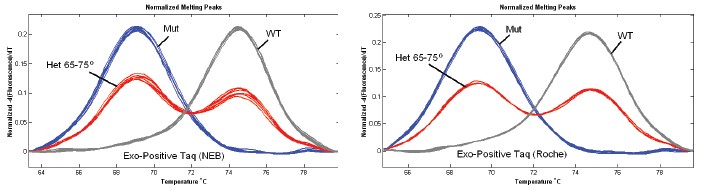
The exonuclease postive (NEB and Roche FastStart) did not show MAAB (65-75 ° C) (Figure 2).
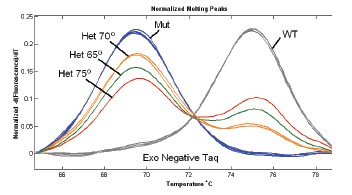
The sensitivity of detecting a mutant allele to each gene of interest in the absence of MAAB using a non-labeled probe was 5-10% (Fig. 3).
When using a hot-start PCR machine for temperature gradients, TP53 x8 and PAH x11 have MAAB (Fig. 4 left). However, malaria-associated genes did not show enhanced mutated alleles on hot-started quantitative PCR instruments (Figure 4 right).
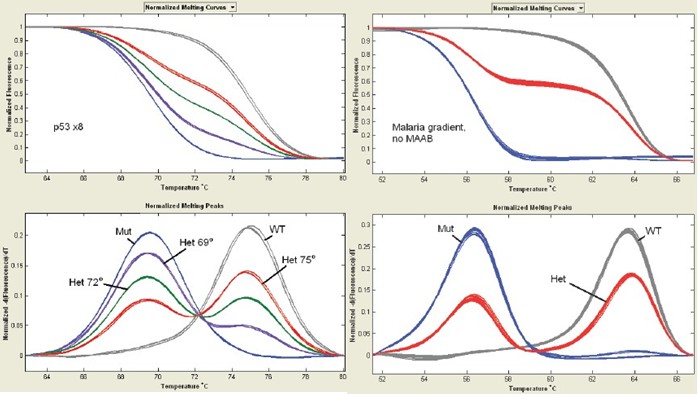
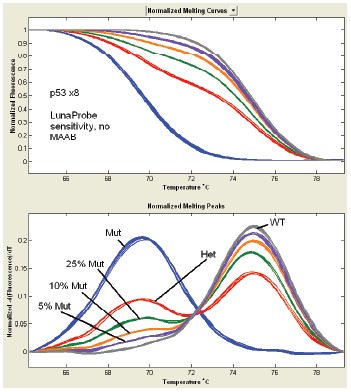
The high-resolution melting curve of the non-labeled probe on the fast cycle PCR instrument and LightScanner32 was then used to perform the bias amplification of the mutant allele using a temperature gradient. Compared to the use of a conventional hot-start PCR machine, the use of a rapid quantitative PCR machine has a significant effect on MAAB at an annealing temperature of 56 °C. The homozygous WT and the mutated allele were mixed (50/50), confirming that the detection rate of the allele of the mutation can be below 1% (Fig. 5) .

Figure 5. Standardized difference peaks for LunaProbes. The wild type allele locus (gray) = 62oC, the mutant allele locus (red) = 54oC. 58oC was used as an annealing temperature to perform differential amplification of mutant alleles of 50–50 mixed samples (yellow). This amplification method allows the resolution of differential amplification to be 10 times and the sensitivity of detection to 0.7–1.5%.
This result confirms that MAAB using unlabeled probes is more sensitive than detection without MAAB.
in conclusion
The following are important parameters for MAAB using unlabeled probes:
1. Temperature conversion rate and temperature of the PCR target slice
2. Annealing temperature and probe Tm value
Exonuclease
1. Temperature conversion rate and temperature of the PCR target slice
2. Annealing temperature and probe Tm value
Exonuclease
After paying attention to these important parameters, the detection rate of the alleles in the study can reach 1%. This method can be used to detect known somatic mutations (p53, EGFR, BRAF) and early detection of parasitic infections. Avoid inappropriate drug treatment and detect fetal DNA mutations with high sensitivity. Quantitative PCR and high-resolution dissolution profile analysis on the LS32, combined with the use of rapid quantitative PCR methods and non-labeled probes are critical for MAAB.
Textile cotton Face Mask is different from disposable face mask. It can be reuseable and promote blood circulation: Wearing a mask in the cold winter can prevent the face from being frostbited, thereby promoting blood circulation on the face
Nonwoven Earloop Face Mask,Earloop Face Mask,5Ply Face Mask,Disposable Mask Template
RFX+CARE Manufacturing Co.,Ltd. , https://www.rk-rfxcare.com