Confirmation process to simplify the results of peptide mapping analysis: a basic function of the Waters Biopharmaceutical System Solution based on UNIFI
The process of confirming the results of the simplified peptide map analysis:
A basic function of the Waters Biopharmaceutical System Solution based on UNIFI
purpose
Demonstrate how the power of the UNIFITM Scientific Information System helps users view and review LC/MS raw data from UPLC®/MS or UPLC/MSE peptide mapping experiments.
background
Peptide mapping is the basic tool for the identification of drug proteins, monitoring new variants of protein drug structures, and monitoring the extent of changes in known variants. Peptide mapping using high-resolution mass spectrometry provides very rich information on protein drug data, and obtaining key data results is very challenging.
Software workflows designed for UPLC/MS peptide mapping, such as the BioPLCLynxTM based on MassLynxTM software and the UPLC/MS peptide mapping process in the UNIFI-based Waters Biopharmaceutical System solution, can significantly reduce data processing time. There is no need to manually view the data set.
However, after the data obtained by the analysts, the analysts often need to re-view the original data of the chromatogram and mass spectrometry to verify the results of the heavy data. Sometimes this is also the requirement of the laboratory SOPs.
The UNIFI Scientific Information System meets the needs of managing biopharmaceutical workflows and has more powerful features to support the user's validation process.
It simplifies the verification process of results, standardizes laboratory processes, and reduces the time from data collection to product decision making.
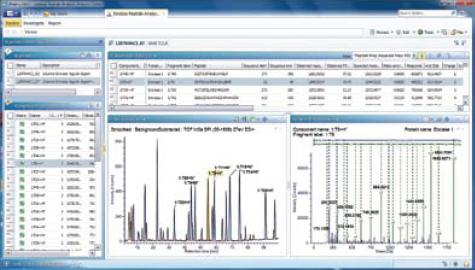
Figure 1. The Review function option in UNIFI allows you to view information about a peptide in the processed UPLC/MSE peptide map data.
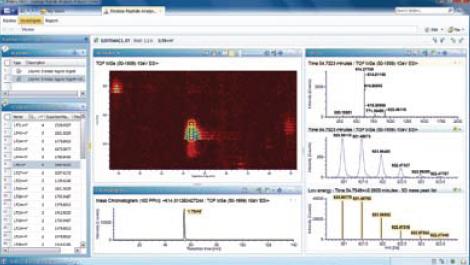
Figure 2. The Investigate function option in UNIFI allows you to view information about each peptide in the UPLC/MSE raw data, as well as open analysis.
The solution collected UPLC/MSE peptide map data (120 minutes, gradient elution) of protein enolase I (from S. cerevisiae). Data processing using the UNIFI UPLC/MS peptide mapping workflow, using typical parameter settings (<10 ppm mass accuracy, trypsin, semi-trypsin and single miscut bias peptide assignment, may result in asparagine deamidation and methionine oxidation) ).
Figure-1 on the previous page is the result of the processed peptide map, which is displayed as a screen shot in the UNIFI Review function option. The Review option provides interactive navigation of the main data tables and the results of processing between different graphical display windows. In the figure, the T6 trypsin hydrolyzed peptide is highlighted in the peak intensity (BPI) spectrum in the chromatogram window (retention time is about 54.75 min), and the relevant MSE mass spectrometry data is displayed in the Fragment Viewer window. This information and the results of the form can be used to quickly determine if the resulting automatic peak assignment is correct.
For some unexpected results, the user can further mine the raw data to verify the quantitative and qualitative analysis results obtained. Open the Investigate Features tab (see Figure 2 on the previous page) to display graphical and tabular display areas of chromatographic and mass spectrometric results directly generated from UPLC/MS raw data.
Figure-1 on the previous page is the result of the processed peptide map, which is displayed as a screen shot in the UNIFI Review function option. The Review option provides interactive navigation of the main data tables and the results of processing between different graphical display windows. In the figure, the T6 trypsin hydrolyzed peptide is highlighted in the peak intensity (BPI) spectrum in the chromatogram window (retention time is about 54.75 min), and the relevant MSE mass spectrometry data is displayed in the Fragment Viewer window. This information and the results of the form can be used to quickly determine if the resulting automatic peak assignment is correct.
For some unexpected results, the user can further mine the raw data to verify the quantitative and qualitative analysis results obtained. Open the Investigate Features tab (see Figure 2 on the previous page) to display graphical and tabular display areas of chromatographic and mass spectrometric results directly generated from UPLC/MS raw data.
These views also allow users to conduct open research, such as extracting peaks of certain qualities, adding spectra, and applying background corrections. Conversely, the corresponding raw data can be analyzed by a certain peak after processing. To simplify the diagram, the same T6 peptide was selected for further study.
The 3D graphical browsing tool in Figure 2 (upper left, 2D view, 2+ ion smart zoom) indicates that there is no interference in the mass spectral data of this peak, and the isotope peak detection results of 2+ ions are highlighted.
More importantly, the T6-related extracted ion chromatogram (bottom left), summation map (upper right, middle right), and processed mass spectrometry data (bottom right) can be automatically selected by using selectable commands related to the composition window. Generated (Figure-3).
The ability to analyze individual peaks in UPLC/MS raw data not only shortens the time for users to view other maps for verification of results, but also standardizes the process of peptide mapping analysis in the laboratory.
More importantly, the T6-related extracted ion chromatogram (bottom left), summation map (upper right, middle right), and processed mass spectrometry data (bottom right) can be automatically selected by using selectable commands related to the composition window. Generated (Figure-3).
The ability to analyze individual peaks in UPLC/MS raw data not only shortens the time for users to view other maps for verification of results, but also standardizes the process of peptide mapping analysis in the laboratory.
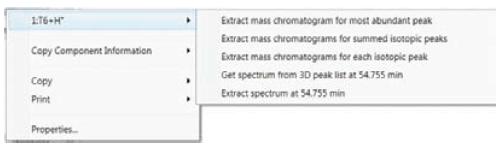
Figure 3. For each detected peak, the extracted mass spectrum, the sum spectrum, and the associated processed mass spectrum are automatically generated.
Summary The ability of scientists to view and reprocess raw data freely under processed data is a basic requirement for UPLC/MS data analysis applications for complex biopharmaceuticals. The UNIFI scientific information system far exceeds this basic requirement by using peptide data detected in peptide mapping to automatically generate a meaningful view of the chromatographic and mass spectral raw data. This platform simplifies the verification process of results, standardizes laboratory processes, and reduces the time from data collection to product decision making.
Ophthalmic Surgical Pack,Disposable Surgical Opthalmic Pack,Surgical Sterile Ophthalmic Pack,Disposable Opthalmic Surgical Pack
Suzhou JaneE Medical Technology Co., Ltd. , https://www.janeemedical.com